
New Phylogenetic Insights on Saitoella Genus from Whole Genome Sequencing of Yeasts Unveiled
Discovery of a New Class in the Saitoella Genus
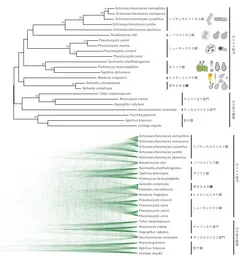
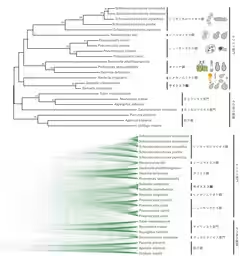
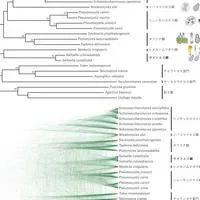
Recent research from the Tokyo University of Agriculture has illuminated the phylogenetic position of the red yeast genus Saitoella, coming 58 years after the initial strain isolation. The study, led by Hiroki Kobayashi and his collaborators, has utilized whole-genome sequencing to perform an exhaustive phylogenetic analysis, leading to the proposal of Saitoellomycetes as a new class within the Taphrinomycotina subphylum.
The primary goal of this research was to decode the genome of two previously unstudied species of the Saitoella genus, particularly Saitoella coloradoensis (the second species in the genus) and Savitreella phatthalungensis from the same subphylum. Through the use of long-read sequencers, researchers managed to connect the entire lengths of each chromosome, providing an unprecedented level of detail in genetic analysis. The results starkly indicate that Saitoella represents a unique lineage distinct from familiar classifications.
Traditionally, yeasts, particularly Saccharomyces cerevisiae, have played critical roles in brewing and baking. The Saitoella genus stands out with pinkish-red colonies, combining morphological features of Basidiomycetes and biochemical characteristics unique to Ascomycetes. Its precise phylogenetic location has remained ambiguous and underexplored until now. As a result of various gene markers previously analyzed, inconsistencies in classification had led to an unclear understanding of Saitoella's taxonomic position.
Upon analyzing the genomes of Saitoella coloradoensis and Savitreella phatthalungensis using advanced sequencing techniques, researchers constructed an entire genome profile that revealed a complex chromosomal structure. Contrary to earlier assumptions, Saitoella's ancestral lineage possesses unambiguous genetic alignments distinct from known groups, suggesting a preservation of ancestral sequences. This was a key factor in the validation of the proposed new class, which is noteworthy due to the unprecedented acknowledgement of a Japanese scientist's name, Dr. Kenji Saito, in the taxonomic hierarchy.
In summary, the genomic analysis showed that Saitoella coloradoensis possesses 38 chromosomes — the highest number observed in known fungi — with an average chromosome size of approximately 370,000 base pairs. The possibility of chromosomal fusion events within closely related species suggests an intriguing evolutionary adaptation. The unique features illuminated through this genomic work raise the potential for industrial applications, particularly in the production of coenzyme Q10 and other valuable compounds.
This notable study, supported by the Fermentation Research Institute, proposes not merely advancement in yeast research but opens the door for exploring the potential of Saitoella in biotechnology. The ramifications of such foundational research extend across various fields, further blending microbiology and industrial applications.
References
- - Kobayashi, H., et al. (2025). Phylogenomic insights into Taphrinomycotina and genomic features of Saitoella coloradoensis based on whole-genome sequencing at the chromosome level, with a proposal of the new class Saitoellomycetes. Mycologia. doi: 10.1080/00275514.2025.2589032.
This groundbreaking study represents a significant leap towards understanding microbial diversity, with implications reaching far beyond academic interest, hinting at practical industrial innovations in the near future.
Topics Other)








【About Using Articles】
You can freely use the title and article content by linking to the page where the article is posted.
※ Images cannot be used.
【About Links】
Links are free to use.