
Innovative Measurement Technique Visualizes Enzyme Dynamics at Atomic Level
Introduction
In all forms of life, including our own bodies, a vast array of molecules interact to sustain a delicate balance of biochemical processes. The proteins known as enzymes act as catalysts in these reactions, controlling how and when molecules interact by binding or cleaving them. Many enzymes undergo structural changes in the order of milliseconds, facilitating a series of processes that include recognizing target molecules, binding, reacting, and releasing products. However, due to their nanoscale size and rapid motion, observing these fine details has been a challenge until now.
This groundbreaking study, led by Mayu Okada and colleagues at Tokyo Metropolitan University, introduces a novel measurement and data analysis technique utilizing Nuclear Magnetic Resonance (NMR) spectroscopy, enabling scientists to visualize the atomic-level structural changes of enzymes. The focus of this research was on the yeast-derived deubiquitinating enzyme YUH1, unveiling how it recognizes and reacts to its target molecule, ubiquitin. The results were published in the Journal of the American Chemical Society on August 7, 2025.
Key Developments
- - The newly developed technique allows for the observation of enzyme structural changes on a millisecond timescale using NMR spectroscopy.
- - By reconstructing the changing structures of enzymes, this research reveals previously hidden mechanisms at play.
- - This exploration delineates how YUH1 dynamically morphs to recognize and process ubiquitin, a fundamental element in cellular protein recycling.
Background
In the human body, various chemical reactions perpetually occur, orchestrated by enzymes that carefully regulate these processes through their inherent structural flexibility. Enzymes facilitate the selection and processing of specific molecules and the release of products post-reaction. This “motion” of enzymes is indispensable for precise chemical modulation. However, capturing such swift, nanoscale changes has traditionally proved difficult.
Prior methods, including X-ray crystallography and cryo-electron microscopy, primarily focused on studying static structures, leaving a gap in understanding the dynamic nature of enzyme behavior during reactions. To address this, the research team developed a novel NMR-based approach aimed at unraveling how enzymes operate while in motion at the atomic level.
Research Details
Using their innovative NMR technique, the researchers meticulously examined the structural dynamics of YUH1. This enzyme plays a critical role in recycling proteins by processing ubiquitin, a molecular marker for proteins slated for degradation. A deeper understanding of how YUH1 interacts with ubiquitin can provide profound insights, especially considering the associations between mutations in the human counterpart of this enzyme, UCHL, with diseases like cancer and Parkinson’s.
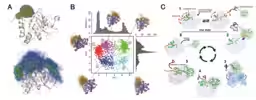
The successful implementation of this technique allowed the team to identify that the end region (N-terminus) and a loop near the active site of YUH1 undergo significant dynamic changes. The N-terminus was found to behave akin to a 'lid', facilitating the entry and exit of ubiquitin as it changed its position in response to its binding. This feature was aptly named ‘gating grid’, indicating its crucial role in substrate recognition.
Tests using enzyme variants that either partially deleted this gating grid or inhibited its motion demonstrated its essentiality for YUH1's enzymatic activity. Binding assays further suggested that the gating grid temporarily interacts with ubiquitin, a new recognition mechanism that provides insight into how enzymes can efficiently seize their substrates.
Significance and Implications
The implications of this research are significant; it establishes a new framework for understanding enzyme dynamics at an unprecedented resolution. This understanding not only deepens our fundamental insights into enzymatic activity but also heralds new avenues for drug discovery and disease treatment targeting various proteins and enzymes.
In summary, this innovative NMR analysis method is anticipated to be applicable to a variety of enzymes and flexible proteins, potentially unraveling the intricate relationships between dynamic structures and biological functions across many life phenomena.
Conclusion
This study marks a pivotal advancement in our comprehension of biochemical processes by illuminating the rapid, intricate dance of enzymes at the atomic scale. With a new lens for observing the relationship between structure and function, researchers can expect breakthroughs in both basic science and the development of therapies for diseases linked to enzymatic dysfunction.
References
- - Mayu Okada, Yutaka Tateishi, Eri Nojiri, Tsutomu Mikawa, Sundaresan Rajesh, Hiroki Ogasa, Takumi Ueda, Hiromasa Yagi, Toshiyuki Kohno, Takanori Kigawa, Ichio Shimada, Peter Güntert, Yutaka Ito, Teppei Ikeya. Multi-state structure determination and dynamics analysis reveals a unique ubiquitin-recognition mechanism in ubiquitin C-terminal hydrolase. Journal of the American Chemical Society. DOI: 10.1021/jacs.5c06502.
Topics Health)










【About Using Articles】
You can freely use the title and article content by linking to the page where the article is posted.
※ Images cannot be used.
【About Links】
Links are free to use.