
Revolutionary Minimal DNAzyme: Catalyzing RNA Cleavage with Just Two Bases
The Development of the World's Smallest DNAzyme
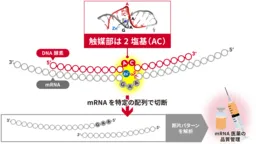
Researchers at the National Institute of Advanced Industrial Science and Technology in Japan have made significant progress in the field of molecular biology by developing the world's smallest DNA enzyme, capable of catalyzing RNA cleavage using only a two-base sequence. This DNAzyme, identified as minGAA, utilizes zinc ions to facilitate the hydrolysis of RNA, offering a groundbreaking tool for quality assessment of mRNA used in vaccines.
Background of the Research
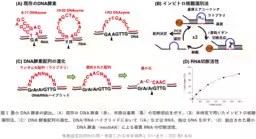
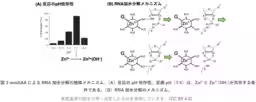
Led by Kazuhiko Yamazaki, a senior researcher in the Molecular Biosystems Research Institute, along with Tohmi Kubota, Shun Miyagishi, Rika Inomata, and others, this project marks a collaboration with experts from the University of Tokyo and Kinki University. Previous DNAzymes have required between ten to fifteen bases to function, but through innovative in vitro selection methods, the team produced a DNAzyme that only needs two bases to serve as its catalytic site, making it the smallest of its kind ever created.
Mechanism of RNA Hydrolysis
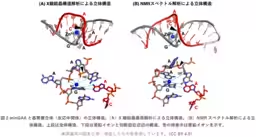
Their comprehensive analysis using X-ray crystallography and NMR spectroscopy shed light on the catalytic mechanism involving the DNAzyme and zinc ions. It was determined that a specific configuration of the zinc ion is crucial for the enzymatic reaction, as it allows the DNAzyme to bind to the RNA substrate effectively. The results mean that the proposed binding sites can target RNA sections containing the specific sequences necessary for the desired cleavage.
Applications in mRNA Vaccine Quality Control
The DNAzyme minGAA holds the promise of being an invaluable asset in the realm of mRNA therapeutics. With mRNA vaccines becoming a focal point in medical advancement, the ability to assess mRNA quality is critical. Traditional sequencing methods help analyze long mRNA sequences but are often time-consuming and not feasible for routine quality control. In contrast, the minGAA DNAzyme can cleave specific RNA segments, thus making it easier to ascertain the presence of any irregularities or structural abnormalities in the mRNA strands by analyzing the cleavage patterns.
Catalytic Characteristics of minGAA
The minGAA DNAzyme exhibits peak activity at pH 7.5, where it efficiently catalyzes nucleic acid cleavage in the presence of zinc. The catalytic mechanism operates through the interaction of the zinc ion with the RNA's guanine base, facilitating the transfer of a negative charge that promotes the cleavage of RNA. Through mass spectrometry, this catalytic outcome can be verified, adding another layer of affirmation to the DNAzyme's functionality.
Future Directions
Moving forward, this research emphasizes the continued development of functional nucleic acid molecules along with further structural analysis to unravel more molecular mechanisms. The success of minGAA not only sets a precedent for future DNAzymes but also revolutionizes how RNA-based therapeutics are evaluated and administered.
The complete findings from this study were published on January 15, 2025, in Nucleic Acids Research, marking a new era in nucleic acid research.
Conclusion
With DNAzymes like minGAA, the future of mRNA therapeutics looks promising. These advancements in enzyme technology will likely play a critical role in the quality management of mRNA vaccines, ensuring safety and efficacy in therapeutic applications. The innovations presented in this study encapsulate a monumental leap in biotechnological applications and present an essential tool for researchers and pharmaceutical developers alike.
Topics Consumer Products & Retail)










【About Using Articles】
You can freely use the title and article content by linking to the page where the article is posted.
※ Images cannot be used.
【About Links】
Links are free to use.