
Introducing AutoFixMark: A Novel Tool for Predicting CO₂ Fixation Pathways in Bacteria
Introduction to AutoFixMark
Researchers from multiple institutions, including the National Institute of Genetics and the National Institute of Advanced Industrial Science and Technology (AIST), have collaborated to develop "AutoFixMark," a sophisticated software tool that predicts the availability of CO₂ fixation pathways in chemolithoautotrophic bacteria from genomic information. This groundbreaking work is crucial in addressing challenges associated with CO₂ utilization and carbon cycling on a global scale.
Background
CO₂ fixation by microorganisms is essential for their survival in carbon-restricted environments and plays a vital role in the Earth's carbon cycle. Chemolithoautotrophic bacteria possess a variety of CO₂ fixation pathways, with seven types, including the well-known Calvin-Benson cycle, documented so far. However, predicting these pathways based solely on genomic information has proven challenging due to the diverse nature of the enzyme genes involved in each pathway. Existing metabolic pathway prediction tools often struggle with the accuracy required to identify these pathways, particularly the recently discovered ones.
Innovations of AutoFixMark
To tackle these challenges, the research team accomplished three key goals:
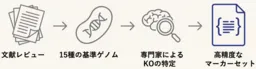
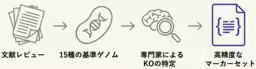
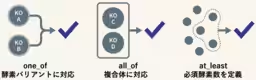
1. Defining Pathway-Specific Marker Genes: The team established essential marker enzymes and their corresponding KEGG Orthology (KO) IDs required to identify each of the seven known CO₂ fixation pathways. This was based on genomic data from 15 representative chemolithoautotrophic bacteria.
2. Developing the AutoFixMark Tool: Using the identified marker enzymes and flexible logical rules for prediction, the team created the AutoFixMark tool. This software can automatically determine the presence of CO₂ fixation pathways based on a gene list derived from genomic data. It operates on Python and is publicly available on GitHub.
3. Establishing a High-Quality Benchmark Dataset: To accurately evaluate the performance of AutoFixMark, a curated reference dataset comprising the genomes of 347 microbial strains, which had verified CO₂ fixation capabilities, was compiled. This dataset enables a precise assessment of AutoFixMark against existing tools like METABOLIC and gapseq, demonstrating superior predictive outcomes, particularly for complex pathways that were previously challenging to assess.
Future Prospects
AutoFixMark opens new avenues for biotechnological applications by facilitating the exploration of microorganisms capable of utilizing CO₂ as a resource. This tool can enhance understanding of the systematic diversity of chemolithoautotrophic bacteria in the environment and support efforts towards a decarbonized society through its application in metagenomic analyses and single-cell genomics. Access to all curated enzymatic gene sets, prediction rules, AutoFixMark software, and the benchmark dataset is freely available, promoting global research collaboration and advancement.
Conclusion
The AutoFixMark tool exemplifies a significant advancement in microbial genomics, providing researchers with a user-friendly platform to explore CO₂ fixation pathways effectively. This pioneering software represents an essential resource in the ongoing pursuit of understanding microbial roles in carbon cycling and the development of sustainable biotechnological solutions.
References
1. Kawashima, S. et al., "A curated resource of chemolithoautotrophic genomes and marker genes for CO₂ fixation pathway prediction," Scientific Data, February 11, 2026. DOI: 10.1038/s41597-026-06655-z
2. AutoFixMark Software: GitHub Repository
3. KEGG Orthology Database.
Glossary
- - Chemolithoautotrophs: Bacteria that convert CO₂ into organic matter using the chemical energy from inorganic substances, rather than light energy.
- - KEGG Orthology: A database categorizing functions of genes and proteins, widely used for functional estimation in genomic analyses.
- - CO₂ Fixation Pathways: Pathways utilized by chemolithoautotrophic bacteria to convert carbon dioxide into organic compounds, including the Calvin-Benson cycle and others.

Topics Other)










【About Using Articles】
You can freely use the title and article content by linking to the page where the article is posted.
※ Images cannot be used.
【About Links】
Links are free to use.