
Revolutionary Machine Learning Method Doubles Crystal Structure Prediction Accuracy
A Major Advancement in Crystal Structure Prediction
Recent research led by Takuya Taniguchi, an associate professor at Waseda University, has achieved a remarkable boost in the success rate of predicting organic molecular crystal structures. Utilizing a new method called SPaDe-CSP, this technique employs machine learning to narrow down the search space for potential crystal structures effectively. This advancement is crucial for the development of pharmaceuticals and functional organic materials, where precise control over crystal structures is essential for enhancing solubility and stability.
Introducing SPaDe-CSP
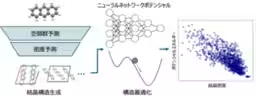
The innovative method called SPaDe, short for Space group and Packing Density predictor, smartly combines machine learning with a neural network potential. This approach allows researchers to filter the most promising space groups and crystal densities early in the prediction process. After this initial filtering, a high-speed and high-accuracy neural network potential is used to optimize the crystal structures, dramatically increasing search efficiency and cutting down on computational costs.
Achievements and Success Rates
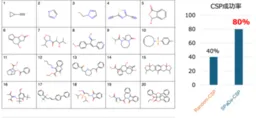
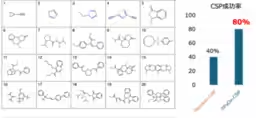
Through experiments conducted on 20 types of organic crystals, the research team successfully predicted the crystal structures with an impressive 80% success rate. This performance signifies a twofold increase compared to traditional random search methods. The implications of this result suggest that SPaDe-CSP can accelerate the discovery of new organic materials, significantly impacting the fields of drug development and electronic materials.
The Importance of Accurate Crystal Structure Prediction
The ability to accurately predict organic molecular crystal structures is of utmost importance for various applications. The structure of these crystals directly influences the properties of pharmaceutical compounds, including solubility and stability, as well as the electrical conductivity of organic semiconductors. Historically, this had posed a significant challenge due to the considerable computational costs involved in evaluating a vast number of possible structures generated via traditional prediction methods.
Expanding Horizons: Future Applications
This new methodology also carries potential for addressing various challenges in drug development and material design. By employing SPaDe-CSP, researchers can now pre-evaluate the risks associated with undesirable polymorphs appearing during the manufacturing process, enabling the selection of the most stable and effective crystal structures. Furthermore, for fields like organic electronics, where molecular arrangement dictates performance, this predictive technique offers a pathway to theoretically explore and design crystal structures exhibiting desired features.
Challenges Ahead
While the introduction of SPaDe-CSP presents significant advantages, there are still areas that require further development. The current assumption that molecular shapes are rigid could limit its application, particularly for flexible molecules that might undergo various conformations during the crystallization process. Future research must address these complexities, incorporating molecular flexibility into the predictive workflow.
Moreover, as the predictions are currently based on the stability assessment at absolute zero degrees Kelvin (0 K), it is also essential to account for temperature effects to evaluate stability at practical operating temperatures, thus enhancing the method's real-world applicability.
Conclusion
The collaborative efforts of the research group at Waseda University demonstrate that the integration of machine learning with neural network potential has the power to transform the field of crystal structure prediction. Given the promising results, expectations are high that this innovation will lead to breakthroughs in the creation of new drugs and innovative materials. With further advancements expected, the goal is to extend the capabilities of SPaDe-CSP to accommodate more complex molecules, paving the way for even broader applications in material science.
Topics Consumer Technology)










【About Using Articles】
You can freely use the title and article content by linking to the page where the article is posted.
※ Images cannot be used.
【About Links】
Links are free to use.